Processing the time-lapse images
05 Jun 2015Extracting timestamps from a time lapse video
This post will outline the workflow I created to extract time-stamps from a time-lapse video. The time-lapse camera I have been using has very little flexibility in setting the the play back frame rate of the stored video. In addition, I wanted to remove all the night time images except for one frame, so the transition from one day to another is obvious.
The way I have approached this was to;
- Use imagemagick to extract all of the frames as jpeg images from the time-lapse video.
- Extract the date characters from each image and use an artificial neural network to classify each character to determine the time of each image.
- Use a second neural network to determine if the image was taken at night (dark) or during the day (light).
- Use ffmpeg to stitch the images back together at my desired frame rate.
Load and examining the training dataset
The first step requires the use of a training set of all the characters to build train a neural network model. Therefore the first set of images must be loaded into R (I manually cropped them before this).
# Load the jpeg library
library(jpeg)
# Load the training set - rounding each pixel off to 0 or 1.
number_zero <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_zero.jpeg"
)
)
number_one <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_one.jpeg"
)
)
number_two <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_two.jpeg"
)
)
number_three <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_three.jpeg"
)
)
number_four <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_four.jpeg"
)
)
number_five <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_five.jpeg"
)
)
number_six <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_six.jpeg"
)
)
number_seven <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_seven.jpeg"
)
)
number_eight <- round(readJPEG(
"_Rmd/training_set/number_wang/number_eight.jpeg"
)
)
number_nine <- round(
readJPEG(
"_Rmd/training_set/number_wang/number_nine.jpeg"
)
)
slash <- round(
readJPEG(
"_Rmd/training_set/number_wang/slash.jpeg"
)
)
dots <- round(
readJPEG(
"_Rmd/training_set/number_wang/dots.jpeg"
)
)
# Combine all of teh examples into a single matrix
covars <- rbind(
matrix(dots, nrow=1),
matrix(slash, nrow=1),
matrix(number_zero, nrow=1),
matrix(number_one, nrow=1),
matrix(number_two, nrow=1),
matrix(number_three, nrow=1),
matrix(number_four, nrow=1),
matrix(number_five, nrow=1),
matrix(number_six, nrow=1),
matrix(number_seven, nrow=1),
matrix(number_eight, nrow=1),
matrix(number_nine, nrow=1))Have a look at the training set using ggplot2
# Load ggplot2 and ggthemes
library(ggplot2)
library(ggthemes)
# Re-format the trainset matrix into a ggplot2 friendly data frame
image_df <- data.frame(x=rep(rep(1:14, each=14), 12),
y=rep(rep(14:1, 14), 12),
z=matrix(sapply(1:dim(covars)[1], function(x) covars[x,])),
character = rep(c(":", "/", 0:9), each = 196))
ggplot() + geom_raster(aes(x,y,fill=factor(z)),image_df) + facet_wrap(~character) +
theme_few(15) + scale_fill_manual(guide = "none",values = c("black", "white"))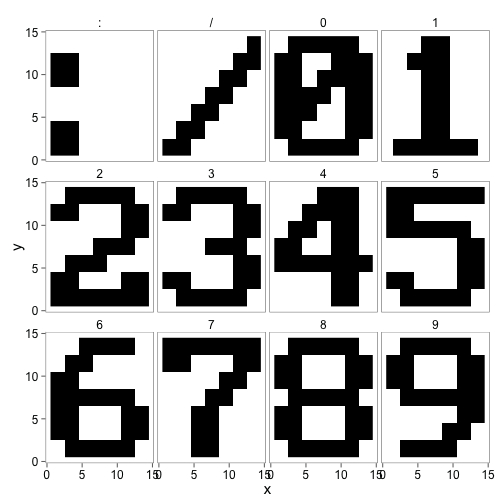
Fit the first neural network
The next section will fit a neural network to the training matrix from above. The training set in this case is the covariates for the neural network, where each row of the covariate matrix contains the pixel values for each character and each column of the covariate matrix representing the value of a certain pixel (e.g. (1,1) of each character). In addition the nnet function needs to know which row of the training set represents which character (the y variable).
# Create a vector with the factors of each row in the training set
timestamp_values <- as.factor(c(":", "/",0,1,2,3,4,5,6,7,8,9))
library(nnet)
# Create a matrix with the correct result labeled in the matrix
nums <- class.ind(timestamp_values)
# Fit the model.
set.seed(1750)
mod <- nnet(x=covars, y = nums,
size = 4,
maxit = 20000,
rang = 0.1,
decay = 5e-4,
trace = FALSE)An example of extracting the time-stamp
This next section will outline how to use the neural network to extract the time-stamp from the original frame of the time lapse images. The original image for this example is:
Firstly, two helper functions are required. The first of these functions uses imagemagick to crop the time-stamp from the bottom of the image and convert it to grey-scale.
# This function creates a black and white image and crops the image
# to the timestamp (manually).
extract_datestamp <- function(filename){
system(paste0("convert -negate -type Grayscale -threshold 80% ",
filename," -gravity South -crop 320x14+53+2 tmp.jpeg" ))
unknown <- round(readJPEG("tmp.jpeg"))
unlink("tmp.jpeg")
return(unknown)
}The next function finds and extracts the characters from the cropped image above and returns the pixels of each character in a matrix with each row being each individual character. This function will go across the image from left to right. It will look at each column of pixels and determine the start of a character when one or more pixels in the column are white, and the end of that character when all pixels are black.
find_characters <- function(x, ncol, nrow ){
character_list <- list()
index <- 1
i = 1
while(i < ncol(x)){
if(sum(x[,i])<nrow(x)){
tmp <- matrix(1, nrow = 1, ncol=ncol*nrow)
j=1
while(sum(x[,i])<nrow(x)){
tmp[j:(j+ncol-1)] <- x[,i]
j = j+ncol
i = i+1
}
character_list[[index]] <- tmp
index = index+1
}
i = i+1
}
character_matrix <- do.call("rbind",character_list)
return(character_matrix)
}And finally the interesting part extracting the time-stamp from the jpeg image. Here is what image looks like after cropping and converting to grey-scale using imagemagick.
# Get the filename
file_to_test <- list.files("_Rmd/training_set/example")
# Use the helper function to extract the timstamp from the image
unknown <- extract_datestamp(paste0("_Rmd/training_set/example/",file_to_test))
# Format the matrix to create an image with ggplot2
image_df <- data.frame(x=rep(14:1, each=320),
y=rep(1:320, 14),
z=matrix(sapply(1:dim(unknown)[1], function(x) unknown[x,])))
# Plot up the timestamp to have a look at it.
ggplot() + geom_raster(aes(y,x,fill=factor(z)),image_df) +
theme_few(15) + scale_fill_manual(guide = "none",values = c("black", "white"))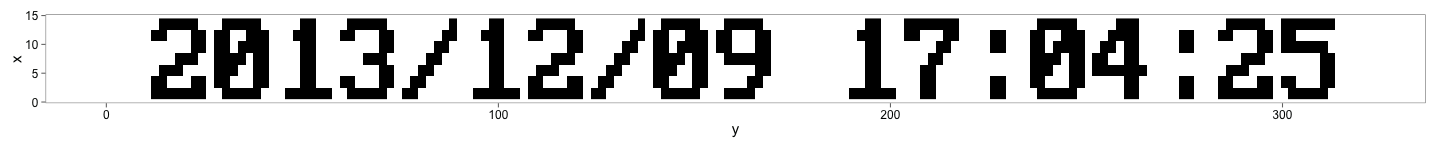
# Build a matrix of the characeters in the timestamp.
characters <- find_characters(unknown, 14,14)
# Predict each of the characters
predicted_characters <- levels(timestamp_values)[max.col(predict(mod, characters))]And now the lubridate package can be used to put the characters together and format them as a date object. Did it get it right?
library(lubridate)
date <- ymd_hms(paste(predicted_characters, collapse=""))
date## [1] "2013-12-09 17:04:25 UTC"Predicting night or day
The next stage of the image processing is to determine if the image is taken during the day or at night. The easiest solution I could think of to solve this was to shrink the larger image down to a workable size (2% of the original size) and fit a new neural network model to predict if the image is dark or not.
The get_sample function uses imagemagick to rescale and convert the image to grey-scale and load it in to R as a matrix.
# This function shrinks the image, converts it to black and white and loads
# it in as a matrix.
get_sample <- function(filename){
system(paste0("convert -negate -resize 2% -type Grayscale ",filename," tmp.jpeg" ))
large <- readJPEG("tmp.jpeg")
unlink("tmp.jpeg")
matrix(large, nrow = 1)
}Now load in the training set. I have pre-selected several dark and light images to use to train the neural network.
# load the daytime images first.
day_files <- list.files("_Rmd/training_set/day_night/day_examples")
day_examples <- matrix(NA, length(day_files),364)
for(i in 1:length(day_files)){
day_examples[i,] <- get_sample(paste0("_Rmd/training_set/day_night/day_examples/",
day_files[i]))
}
# Now load in the night time images
night_files <- list.files("_Rmd/training_set/day_night/night_examples")
night_examples <- matrix(NA, length(night_files),364)
for(i in 1:length(night_files)){
night_examples[i,] <- get_sample(paste0("_Rmd/training_set/day_night/night_examples/",
night_files[i]))
}
# Combine the images together into one larger matrix.
nd_examples <- rbind(day_examples, night_examples)Like earlier, the model is now built based on this training set with each row representing all the pixels from one image.
## Build a model to predict night and day
values <- as.factor(c(rep("day", length(day_files)), rep("night", length(night_files))))
nums <- class.ind(values)
library(nnet)
night_day_mod <- nnet(x=nd_examples,
y = nums,
size = 2,
maxit = 20000,
rang = 0.1,
decay = 5e-4,
trace = FALSE)An example using the model to classify the image
Using the example image from earlier (this is in the training set) here is an example of what is going on.
# Get the filename
file_to_test <- list.files("_Rmd/training_set/example")
# Use the helper function to extract the timstamp from the image
unknown <- get_sample(paste0("_Rmd/training_set/example/",file_to_test))
# Format the matrix to create an image with ggplot2
image_df <- data.frame(x=rep(14:1, 26),
y=rep(1:26, each=14),
z=as.numeric(unknown))
# Plot up the timestamp to have a look at it.
ggplot() + geom_raster(aes(y,x,fill=z),image_df) +
theme_few() + scale_fill_gradient(guide="none",low = "#f0f0f0", high = "#636363")
# And now to predict if the image was taken during the day or at night.
# The predition is the probablity of the image having been taken during the day or at night.
predict(night_day_mod, get_sample(paste0("_Rmd/training_set/example/",file_to_test)))## day night
## [1,] 0.9889661 0.01103407Putting it all together
This section will outline how to use the models developed earlier to extract the images from multiple videos. The code extracts the images, classifies them, removes the unwanted images and stitches the images together using the desired frame rate.
# Create some directories to store the extracted image from.
# Move across to the folder with the time lapse videos
setwd("~/Desktop/timelapse/working")
# create a directory to store the images in
dir.create("images")
dir.create("finished")
# get the file names of the videos to process.
files_to_process <- dir(pattern=".AVI")
# The next part is a little long winded as it contains
# a two nested for loops. Explanations can e found within the code below.
# Loop over the video files
for(i in 1:length(files_to_process)){
# Extract all of the image from the timelapse video and place them into
# the images directory
system(paste0("ffmpeg -i ",
files_to_process[i]," -f image2 -q:v 1 images/tmp-%03d.jpeg"))
# Get a list of the images which were just extracted
images_to_process <- dir("images",pattern=".jpeg")
# Create a list to store the processed dates in
dates <- list()
# Create a list to store the results of the day time or
# night time classification
is_dark <- list()
# Now loop over each image and classify them one by one.
for(j in 1:length(images_to_process)){
# Clip the image to get the timestamp part of the image
datestamp <- extract_datestamp(paste0("images/",images_to_process[j]))
# Extract the characters from the timestamp
characters <- find_characters(datestamp, 14,14)
# Predict each of the characters
predicted_characters <- levels(timestamp_values)[max.col(
predict(mod, characters))
]
# Format the date and add it to the date list
dates[[j]] <- ymd_hms(paste(as.character(predicted_characters),
sep="", collapse = ""))
# See if the photo was taken at night
is_dark[[j]] <- max.col(predict(
night_day_mod,
get_sample(paste0("images/",images_to_process[j]))))==2
}
# Now convert the dates and the dark list to vectors.
dates <- do.call("c", dates)
is_dark <- do.call("c", is_dark)
dark_images <- !is_dark
# Find which image was taken between 12 and 1 am.
midnight_image <- hour(dates)==0
# Create a vector to remove all night time images except the midnight image.
remove_index <- (dark_images+midnight_image)==0
# Remove the nigh time images.
unlink(paste0("images/",images_to_process[remove_index]))
# For the re-stitching back to a video ffmpeg requires the filenames to be incremental.
# This next part renames all of the files in the correct order
new_names <- paste0("tmp-",
sprintf("%03d",
1:length(images_to_process[!remove_index])), ".jpeg")
for(j in 1:length(new_names)){
system(paste0("mv images/",
images_to_process[!remove_index][j],
" finished/", new_names[j]))
}
# And now put create the new video from the processed images.
system(
paste0("ffmpeg -f image2 -r 5 -i finished/tmp-%03d.jpeg -y -r 25 -q:v 1 final_",
i,".mpg"))
}
# Now once the videos have been processed remove all the junk
unlink("finished", recursive = TRUE)
unlink("images", recursive = TRUE)